This Argonne biochemist works on developing biochips, which can identify proteins and used to trace the origins of biological agents such as anthrax in addition to their ability to diagnose infections and even cancer.
June 30, 2011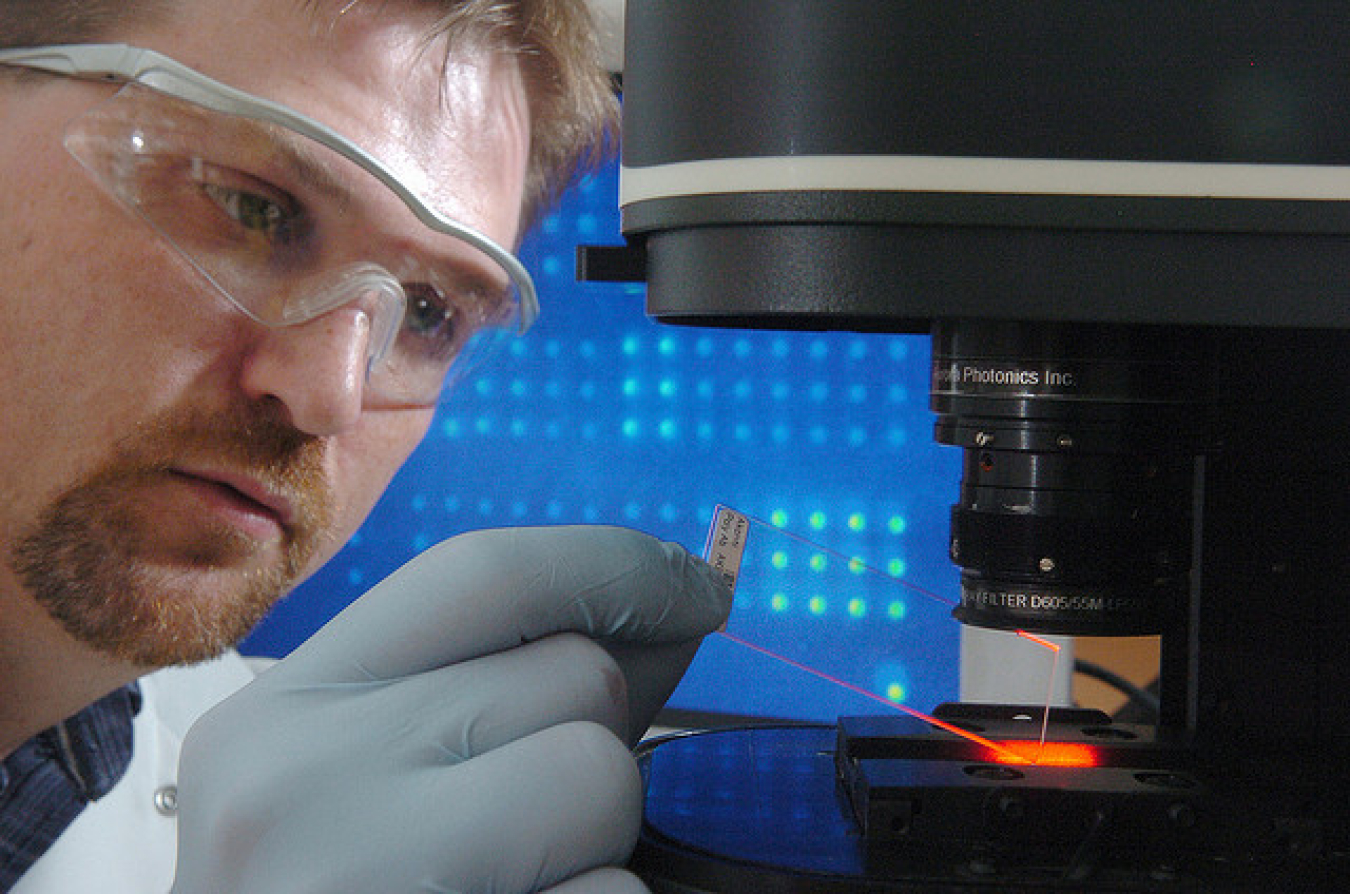
Biochemist Dan Schabacker uses microbial forensics technology -- using proteins in microbes to get clues about how they were created -- to detect where organisms, such as anthrax spores, came from. | Courtesy of Argonne National Laboratory
Editor’s Note: This is cross-posted from Argonne National Laboratory.
Dan Schabacker is an Argonne biochemist who works on developing biochips, tiny arrays that identify proteins that can be used to trace the origins of biological agents such as anthrax. Biochips could also be used as diagnostic medical tools for infections and even cancer. Question: How did you first get interested in science? Dan Schabacker: I was always interested in biology, from grade school through high school. I didn't know what I wanted to do, but I liked life sciences. So I went to the University of Illinois and majored in biology, and that's where I got my more specific interest in immunology. Basically, that's studying how the human immune system responds to attacks. In grad school, my research attempted to exploit the body's immune system to prevent cocaine from crossing the blood-brain barrier -- in essence developing a vaccine to block the euphoric, addictive nature of cocaine. I was always very practical. Although we certainly need basic science to build on, my particular interest has always been in applied science. So in my Ph.D. program, when we had developed a prototype vaccine for cocaine addiction, we actually made something useful. I liked that. Q: What are you working on now? DS: We've switched over the last two years from biodetection -- which everyone can do, it's a very established science now -- to what we call characterization. So not only can we detect biological terror agents, but we can provide information about the organism -- where it was from originally. That's the forensics we're doing now. Additionally our signal development approach is unique because we use slightly different methods. Most of the current devices in the diagnostic community are fluorescent-based; they use light or lasers and optics to capture an image. But we're looking at systems that don't use optics; instead, we use electrochemical types of detection. Basically, our approach allows you to make the device much smaller. The smallest devices using lasers and optics are about the size of a coffee maker. But without the need for that special equipment, you can go smaller, like a hand-held device. Like a glucometer, which is what people with diabetes use to check blood sugar levels -- these are extremely small devices with a disposable strip that you put a drop of blood on, you touch it, it sucks the sample in and tells you what the glucose levels in your blood are. So we want to do the same thing, only with infectious agents. For example, you'd put a respiratory sample on the strip, and it'd tell you -- you have strep throat, you have pneumonia, etc. That's down the road, of course. Q: What attracts you to national security work? DS: One thing I like about national security is the emphasis on having a product at the end. They want an actual widget that can do something, which appeals to me because I like to work on things that have a direct impact. It's also because I think my work can be really helpful to national security. There's a definite need for characterization. There's great interest in attribution -- tracking biological agents to a facility or group or location -- and I believe we can help do that. Q: What's different about your approach? DS: Well, detecting things is relatively well established. But microbial forensics, which is what we do -- using proteins in microbes to get clues about how they were created -- is really new. That field took off after the anthrax scares in 2001; we learned that we need more technology to be able to track the anthrax spores down to where they came from. Most of the field right now is concerned with genetics -- DNA analysis. While genetic analysis can identify the particular strain, it can't tell you how the organism was grown or prepared. Identical strains of anthrax are grown in labs all over the world, so when you are trying to track it down to a specific laboratory or location, knowing the strain may not be enough. Our microbial forensics technology can tell you much more: proteins change according to the lab conditions under which they were grown, for example, which can be very helpful. In general, proteins are much more complex and finicky than DNA, but you can get a lot more -- and different -- information from them. Q: How did you get the idea of using proteins for analysis? DS: The tools that we are developing using proteins actually came out of cancer research. In fact, we are currently collaborating with the University of Chicago on a liver cancer project. The technology used is called a natural proteome biochip -- when I say "proteome," I mean all the proteins expressed by an organism at a given time under specified conditions. We place all the proteins, or the proteome, onto our biochip for characterization. In the case of cancer, we take samples of the tumor and get a "signature" of all the proteins that are expressed by this cancer cell. Then we "interrogate" the signature, looking for differences in protein modification. For example, many times in cancer what you'll see is aberrant glycosylation or phosphorylation patterns --- proteins that are glycosylated or phosphorylated that shouldn't be, or the other way around. So we identify these changes with our tool and get a set of markers indicative of cancer. This could be a diagnostic test: a doctor would test samples from patients to look for these markers. The ultimate goal is to find cancer earlier, because of course the earlier you can find and treat cancer, the better the patient prognosis. We have already observed differences in patients with advanced cancer. In one study we compared serum from patients with advanced melanoma to healthy patients, and were able to use our signature analysis to differentiate one group from the other. We're still working on this project. I believe what is needed are correlation markers: an array of changes that would indicate cancer -- not just a single smoking gun marker. I think that's where cancer diagnostics is headed. That's of course from my limited experience in the field. While working on these cancer projects I thought, why couldn't we use this for infectious diseases? That's how we initiated the forensic-type applications that we're working on now. Q: What do you do for fun when you're not working? DS: My wife Jean and I have three kids: Michelle, 8, Steven, 10 and Chris, 11. My hobby is sports with the kids. I coach every sport you can imagine. Baseball's starting up now, and I also coach basketball and football. (My daughter plays soccer, which I don't coach anymore, I know nothing about soccer.) We're big Illinois fans, too, so we travel to Champaign to watch games. So at this point, there's no time for anything else, but I wouldn't change a thing! Interview by Louise Lerner.
